Verification#
import dataset
import pprint
First we will load Dataset 1 from the paper. Her you have the option to either use the full resolution dataset or a dataset that is downsampled. To speed up computations you can use the downsampled dataset
full_resolution = False
data = dataset.load_sample_data(full_resolution=full_resolution)
pprint.pprint(data.info)
{'dt': 25.004229346742502,
'num_frames': 236,
'size_x': 613,
'size_y': 145,
'time_unit': 'ms',
'um_per_pixel': 1.08615103979884}
We can now extract the first frame from the dataset which will be used in the verification.
frame = data.frames[:, :, 0].T
frame
array([[35754, 36999, 39521, ..., 27215, 22756, 27350],
[36789, 37563, 40376, ..., 27309, 22721, 26453],
[38119, 38877, 41162, ..., 27115, 22545, 27679],
...,
[42218, 43225, 40372, ..., 16339, 15880, 16158],
[38348, 40989, 38138, ..., 16571, 16318, 15839],
[38603, 40368, 38705, ..., 19332, 18579, 17934]], dtype=uint16)
As we can see, the frame is just a regular numpy array with 16 bit unsigned integers.
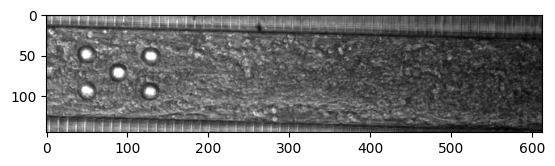
Now let us plot the first frame
import matplotlib.pyplot as plt
plt.imshow(frame, cmap='gray')
<matplotlib.image.AxesImage at 0x7f1190fb6b60>
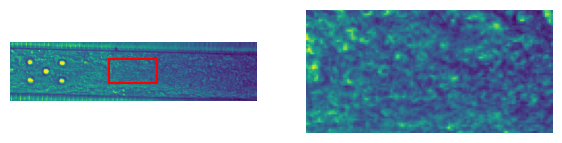
For the verification we will only use a small region of the frame which we can plot as follows
def extract_region(frame, full_resolution):
offset = 100 if full_resolution else 30
n1, n2 = frame.shape
x1 = n1 // 2 - offset
x2 = n1 // 2 + offset
y1 = n2 // 2 - 2 * offset
y2 = n2 // 2 + 2 * offset
return frame[x1:x2, y1:y2]
def plot_frame_with_region(frame, full_resolution):
offset = 100 if full_resolution else 30
fig, ax = plt.subplots(1, 2, figsize=(7, 2))
ax[0].imshow(frame)
n1, n2 = frame.shape
x1 = n1 // 2 - offset
x2 = n1 // 2 + offset
y1 = n2 // 2 - 2 * offset
y2 = n2 // 2 + 2 * offset
ax[0].plot([y1, y2], [x1, x1], "r")
ax[0].plot([y1, y2], [x2, x2], "r")
ax[0].plot([y1, y1], [x1, x2], "r")
ax[0].plot([y2, y2], [x1, x2], "r")
ax[1].imshow(frame[x1:x2, y1:y2])
for axi in ax:
axi.axis("off")
plt.show()
plot_frame_with_region(frame, full_resolution)
region = extract_region(frame, full_resolution)
region
array([[26974, 27902, 31512, ..., 47756, 44681, 34804],
[28237, 28702, 27581, ..., 47156, 46218, 36131],
[28955, 28350, 28022, ..., 52517, 47131, 43947],
...,
[32777, 34007, 32224, ..., 29435, 29171, 30942],
[36765, 33816, 30995, ..., 30707, 33817, 34103],
[43151, 39674, 35979, ..., 34079, 32265, 32381]], dtype=uint16)
We will now go ahead and run the verification using a synthetic function. First we will import some necessary libraries, including mps-motion
from pathlib import Path
from mps_motion import farneback, lucas_kanade, block_matching, dualtvl1
import dask.array as da
from scipy.ndimage import geometric_transform
import numpy as np
import time
import json
Now we will define the synthetic function which is
def get_func(a=0.001, b=0):
def func(x):
return (x[0] * (1 - a), x[1] * (1 - b))
return func
We will use the parameters \((a, b) \in \{ (0.005, 0.0), (0.0, 0.007), (0.005, 0.007) \}\), so we put those into a dictionary
displacement_parameters = {
"x": {"a": 0.005, "b": 0.0},
"y": {"a": 0.0, "b": 0.007},
"xy": {"a": 0.005, "b": 0.007},
}
Now we create new directory where we will store the results
results = Path("results") / "verification"
results.mkdir(exist_ok=True, parents=True)
And then we run the 10 times and code keep the average time
timings = {}
N = 10
for flow, name, kwargs in [
(farneback.flow, "farneback", {}),
(lucas_kanade.flow, "lucas_kanade", {}),
(block_matching.flow, "block_matching", {}),
(dualtvl1.flow, "dualtvl1", {}),
]:
print(name)
timings[name] = {}
for direction in ["x", "y", "xy"]:
path = results / f"{direction}_displacement_{name}.npy"
a = displacement_parameters[direction]["a"]
b = displacement_parameters[direction]["b"]
print(f"{direction} displacement")
frame2 = geometric_transform(region.T, get_func(a, b)).T
t0 = time.perf_counter()
for _ in range(N):
u = flow(frame2, region, **kwargs)
timings[name][direction] = (time.perf_counter() - t0) / N
if isinstance(u, da.Array):
u = u.compute()
np.save(path, u)
path.with_name("timings").with_suffix(".json").write_text(json.dumps(timings))
farneback
x displacement
y displacement
xy displacement
lucas_kanade
x displacement
y displacement
xy displacement
block_matching
x displacement
y displacement
xy displacement
dualtvl1
x displacement
y displacement
xy displacement
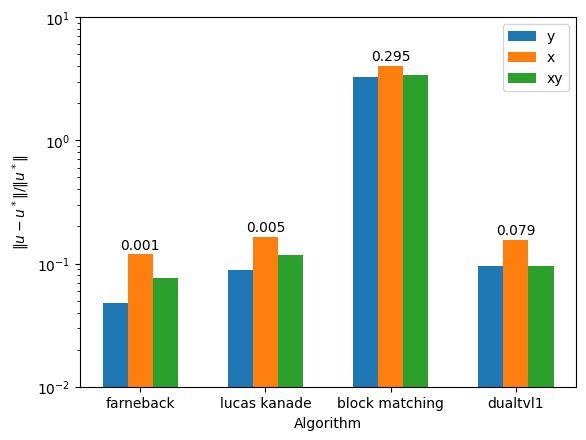
Now we will plot the timings and errors
algs = ["farneback", "lucas_kanade", "block_matching", "dualtvl1"]
cases = ["x", "y", "xy"]
path = results / "timings.json"
timings = json.loads(path.read_text())
x, y = np.meshgrid(np.arange(region.shape[1]), np.arange(region.shape[0]))
all_errors = []
for i, alg in enumerate(algs):
errs = []
for j, disp in enumerate(cases):
U = np.load(results / f"{disp}_displacement_{alg}.npy")
u = U[:, :, 0]
v = U[:, :, 1]
a = displacement_parameters[disp]["a"]
b = displacement_parameters[disp]["b"]
func = get_func(a, b)
u0, v0 = func([x, y])
u_exact = x - u0
v_exact = y - v0
errs.append(
np.linalg.norm([u - u_exact, v - v_exact])
/ np.linalg.norm([u_exact, v_exact])
)
all_errors.append(errs)
all_errors = np.array(all_errors)
width = 0.2
x = np.arange(len(algs))
fig, ax = plt.subplots()
rectsy = ax.bar(
x - width,
all_errors.T[0],
width=width,
align="center",
label="y",
)
rectsx = ax.bar(
x,
all_errors.T[1],
width=width,
align="center",
label="x",
)
rectsxy = ax.bar(
x + width,
all_errors.T[2],
width=width,
align="center",
label="xy",
)
ax.set_xticks(x)
ax.set_xticklabels([" ".join(alg.split("_")) for alg in algs]) # , rotation=30)
ax.legend()
ax.set_xlabel("Algorithm")
ax.set_yscale("log")
ax.set_ylabel(r"$\| u - u^* \| / \| u^* \|$")
for label, rects in [("x", rectsx)]: # , ("y", rectsy), ("xy", rectsxy)]:
for text, rect in zip([timings[alg][label] for alg in algs], rects):
height = rect.get_height()
ax.text(
rect.get_x() + rect.get_width() / 2.0,
1.05 * height,
f"{text:.3f}",
ha="center",
va="bottom",
)
ax.set_ylim(1e-2, 10)
(0.01, 10)
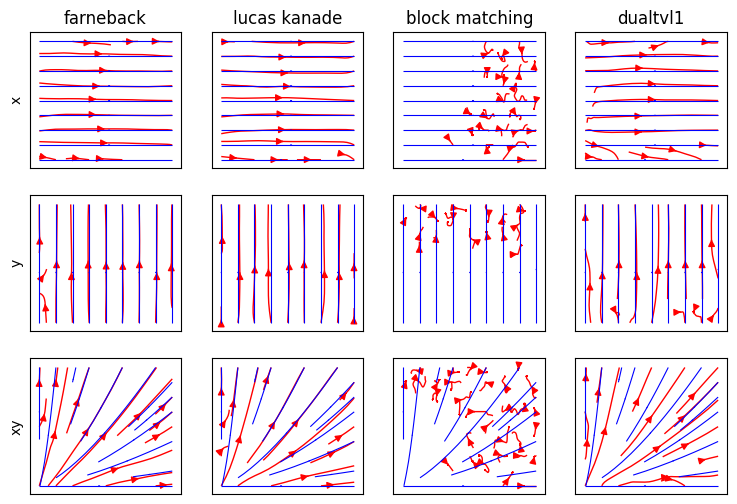
And finally the resulting motion vectors
fig, ax = plt.subplots(3, 4, figsize=(9, 6), sharex=True, sharey=True)
algs = ["farneback", "lucas_kanade", "block_matching", "dualtvl1"]
x, y = np.meshgrid(np.arange(region.shape[1]), np.arange(region.shape[0]))
for i, alg in enumerate(algs):
for j, disp in enumerate(["x", "y", "xy"]):
U = np.load(results / f"{disp}_displacement_{alg}.npy")
u = U[:, :, 0]
v = U[:, :, 1]
a = displacement_parameters[disp]["a"]
b = displacement_parameters[disp]["b"]
func = get_func(a, b)
u0, v0 = func([x, y])
axi = ax[j, i]
axi.streamplot(
x,
y,
u,
v,
color="r",
density=0.3,
linewidth=1.0,
arrowstyle="-|>",
)
axi.streamplot(
x,
y,
x - u0,
y - v0,
color="b",
density=0.3,
linewidth=0.8,
arrowstyle="-",
)
if i == 0:
axi.set_ylabel(disp)
if j == 0:
axi.set_title(" ".join(alg.split("_")))
axi.set_xticks([])
axi.set_yticks([])