Extracting currents#
Sometimes you would like to extract certain intermediate values, that are not state variables from your simulation. One example are the currents in the underlying cellmodel. In this demo we will demonstrate how to extract on particular current (the NaL) current using the built-in fully coupled model.
First we will make the necessary imports.
import simcardems
from pathlib import Path
import dolfin
import matplotlib.pyplot as plt
try:
import ufl_legacy as ufl
except ImportError:
import ufl
Next thing we do is to define the expression for the intermediate that we want to keep track of. In out case we have simple looked at the source code of the cell model and extracted the relevant equations into a function. We have named this function INaL since it will give us an expression for the INaL current. This function takes two arguments; the state vector from the EP solver and the parameters for the ODE model.
def INaL(vs, parameters):
(
v,
CaMKt,
m,
hf,
hs,
j,
hsp,
jp,
mL,
hL,
hLp,
a,
iF,
iS,
ap,
iFp,
iSp,
d,
ff,
fs,
fcaf,
fcas,
jca,
ffp,
fcafp,
nca,
xrf,
xrs,
xs1,
xs2,
xk1,
Jrelnp,
Jrelp,
nai,
nass,
ki,
kss,
cass,
cansr,
cajsr,
XS,
XW,
CaTrpn,
TmB,
Cd,
cai,
) = vs
# Assign parameters
scale_INaL = parameters["scale_INaL"]
nao = parameters["nao"]
F = parameters["F"]
R = parameters["R"]
T = parameters["T"]
CaMKo = parameters["CaMKo"]
KmCaM = parameters["KmCaM"]
KmCaMK = parameters["KmCaMK"]
# Drug factor
scale_drug_INaL = parameters["scale_drug_INaL"]
# Population factors
scale_popu_GNaL = parameters["scale_popu_GNaL"]
HF_scaling_CaMKa = parameters["HF_scaling_CaMKa"]
HF_scaling_GNaL = parameters["HF_scaling_GNaL"]
# Init return args
# Expressions for the CaMKt component
CaMKb = CaMKo * (1.0 - CaMKt) / (1.0 + KmCaM / cass)
CaMKa = (CaMKb + CaMKt) * HF_scaling_CaMKa
# Expressions for the reversal potentials component
ENa = R * T * ufl.ln(nao / nai) / F
# Expressions for the INaL component
GNaL = 0.0075 * scale_INaL * scale_drug_INaL * scale_popu_GNaL * HF_scaling_GNaL
fINaLp = 1.0 / (1.0 + KmCaMK / CaMKa)
return (-ENa + v) * ((1.0 - fINaLp) * hL + fINaLp * hLp) * GNaL * mL
Now we need to opt in to the coupling object to make sure that we registerINaL to the Datacollector. To do so we will use the existing EMCoupling class, but override a few mew methods. We must also remember to call on the super class so that the original methods are invoked.
class EMCoupling(simcardems.models.fully_coupled_ORdmm_Land.EMCoupling):
def __init__(
self,
geometry,
**state_params,
) -> None:
super().__init__(geometry=geometry, **state_params)
self.INaL = dolfin.Function(self.V_ep, name="INaL")
def register_datacollector(self, collector) -> None:
super().register_datacollector(collector=collector)
collector.register("ep", "INaL", self.INaL)
def ep_to_coupling(self):
super().ep_to_coupling()
self.INaL.assign(
dolfin.project(
INaL(
self.ep_solver.vs,
parameters=self.ep_solver.ode_solver._model.parameters(),
),
),
)
We are now ready to run the model. First, we need to load some geometry, and we will use the slab geometry in this case
geo = simcardems.geometry.load_geometry(mesh_path="geometries/slab.h5")
Now, we need to create the custom coupling object. Note however that the CellModel and the ActiveModel remains the same
coupling = simcardems.models.em_model.setup_EM_model(
cls_EMCoupling=EMCoupling,
cls_CellModel=simcardems.models.fully_coupled_ORdmm_Land.CellModel,
cls_ActiveModel=simcardems.models.fully_coupled_ORdmm_Land.ActiveModel,
geometry=geo,
)
We also need to create the configuration, and we pass in the output directory and the coupling_type.
outdir = Path("results_extract_currents")
config = simcardems.Config(
outdir=outdir,
coupling_type=coupling.coupling_type,
)
Now we create a runner for running the simulation
runner = simcardems.Runner.from_models(coupling=coupling, config=config)
And then we run a simulation for 1000 milliseconds
runner.solve(1000)
When the simulation is done we can load the results from the output directory using simcardems.DataLoader
loader = simcardems.DataLoader(outdir / "results.h5")
We can extract the traces from the loader, and specify that the traces we want to extract should be the average over the mesh
values = simcardems.postprocess.extract_traces(loader, reduction="average")
Now we can plot the voltage and the INaL current
fig, ax = plt.subplots(2, 1)
ax[0].plot(values["time"], values["ep"]["V"])
ax[0].set_title("Voltage")
ax[1].plot(values["time"], values["ep"]["INaL"])
ax[1].set_title("INaL")
fig.savefig(outdir / "currents.png")
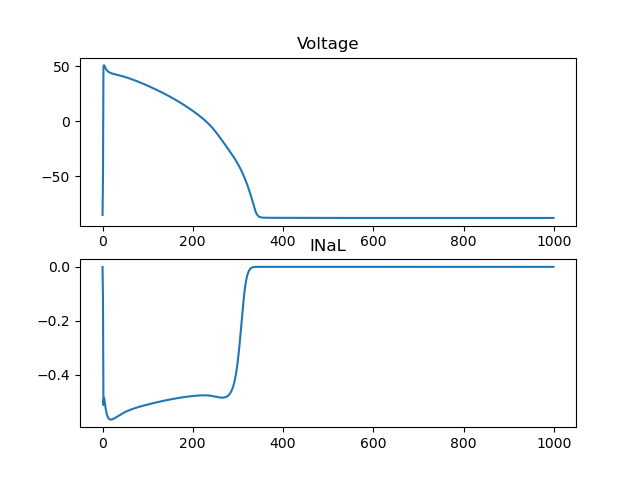
Fig. 8 Showing the voltage and the INaL current#
